 |  |  |
The EVOLUNET project
The EVOLUNET project is supported by the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013 Grant Agreement # 615274, category LS8).
PROJECT MAIN GOALS:
- To introduce and develop sequence similarity networks as a promising complement to the phylogenetic framework to study evolutionary biology.
RATIONALE
The comparison of molecular sequences from organisms, environmental samples and mobile genetic elements is fundamental in evolutionary biology to study genes, genomes and species evolution. Although phylogenetic methods play a major role in these analyses, through the inference of phylogenetic trees or networks based on families of homologous sequences, many data remained underexplored as the number of molecular sequences, especially from microbial genomes and environmental projects, grows exponentially, with typical environmental samples containing thousands of species represented by millions of sequences. In order to further enhance the evolutionary comparison of molecular sequences, we will develop a promising complementary approach, the use of large complex similarity networks (typically a few million nodes and tens of millions of edges), adapted to evolutionary biology questions. Such networks, inspired by the studies on social and regulatory networks, allow for fast inclusive comparative analyses of both (highly) divergent and conserved sequences, fully or partly similar.
We will develop similarity networks to:
- identify and study chimeric genes and genomes,
- detect and analyze the common ‘genetic goods’ shared between gene families and within microbial communities,
- test whether genetic diversity in environmental sequences is significantly larger, both quantitatively and qualitatively, than genetic diversity in some important gene families of cultured organisms (and their mobile genetic elements),
- improve the statistical framework to compare similarity networks.
We expect that our findings will provide an additional comparative framework for evolutionary studies, new methods of detection of environmental/genomic sequences of medical and biological interest, and a novel description of the structure of genetic diversity on Earth, unraveling the main partnerships and barriers to the exchange of sequences of DNA between genes, genomes, species and environments.
LATEST NEWS
15/04/2019: Workshop and hands-on session on networks and evolution in Paris
25/06/2018-29/06/2018: EVOLUNET 2ND SUMMER SCHOOL ON NETWORKS IN ROSCOFF

01/01/18 to 28/02/18: Dr. Diego Javier Zea officially joins the team as a post-doctoral fellow to study the evolution of domain architecture, using networks.
01/12/17 to 31/12/17: Dr. Jananan Pathmanathan works as a post-doctoral fellow for the EVOLUNET project.
04/09/17 to 27/12/17: We welcome Dr. Diego Javier Zea (from the Structural Bioinformatics Units of the Leloir Institute Foundation, Argentina), with whom we will develop networks to study the evolution of protein architectures.

03/05/2017: The Science Breaker highlights our research: « Symbiogenesis: how algae and bacteria shaped new genes together » (https://doi.org/10.25250/thescbr.brk046)
03/05/2017: Mr. Eduard Ocaña (PhD student in the Ruiz-Trillo lab) in training for 3 months in the EVOLUNET team to learn network methods.

15/04/2017: Dr. Guillaume Bernard (post-doctoral fellow) joined the EVOLUNET team. Dr. Guillaume Bernard will develop a statistical framework for network comparisons.

01/10/2016: Dr. Andrew Watson (post-doctoral fellow) and Mr. Romain Lannès (PhD student) joined the EVOLUNET team. Dr. Watson will develop network methods to search for genetic public goods. Mr. Lannes will develop methods to detect divergent environmental genes and microbes.


01/07/2016-31/06/2017: EVOLUNET welcomes Pr. François-Joseph Lapointe (U. Montréal, Canada) for a one year sabbatical in Paris.
04/07/2016-09/07/2016: EVOLUNET 1RST SUMMER SCHOOL ON NETWORKS IN ROSCOFF

18/01/2016: EVOLUNET welcomes its first Master 2 student: Mr. Adrien Danzon, and a visiting scientist: Mr. Alexander Jaffe (visiting scientist). Both investigate environmental genetic diversity using networks.
05/01/2016-09/01/2016: EVOLUNET is honored to welcome Pr. Luay Nakhleh to develop collaborations on sequence similarity networks. To know more about Pr. Nahkleh, check: https://www.cs.rice.edu/~nakhleh/

15/07/2015: EVOLUNET welcomes Mr. Sukithar Kochappi Rajan to start studies of environmental genetic diversity for 4 months.

01/01/2015: Dr. Mattis List, eminent historical linguist, joined the lab to develop network methods, applied to language evolution.
24/11/2014: Dr. Shijulal Nelson-Sathi and Pr. William Martin visit the EVOLUNET team to consider collaborations on reticulate evolution.
01/11/2014: A new post-doctoral fellow, Dr. Eduardo Corel, joins the team EVOLUNET to work on gene and genome networks.

01/07/2014-06/07/2014: The lab welcomes Pr. François-Joseph Lapointe to develop collaborations on sequence similarity networks. To know more about FJL’s multiple talents, check this.
01/06/2014: A post-doctoral position (starting in November 2014) is available. Applications are welcome.
01/06/2014: EVOLUNET welcomes its first PhD student: Mr. Jananan Pathmanathan

01/06/2014: EVOLUNET officially begins.
*PUBLICATIONS*
23.
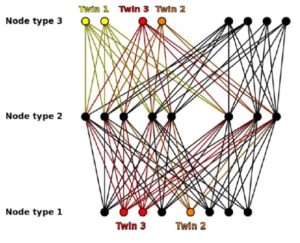
22. Corel E, Pathmanathan JS, Watson AK, Karkar S, Lopez P, Bapteste E. MultiTwin: a software suite to analyze evolution at multiple levels of organization using multipartite graphs. Genome Biol Evol. 2018 Sep 22. doi: 10.1093/gbe/evy209. [Epub ahead of print]
21. Méheust R, Watson AK, Lapointe FJ, Papke RT, Lopez P, Bapteste E. Hundreds of novel composite genes and chimeric genes with bacterial origins contributed to haloarchaeal evolution. Genome Biol. 2018 Jun 7;19(1):75. doi: 10.1186/s13059-018-1454-9.
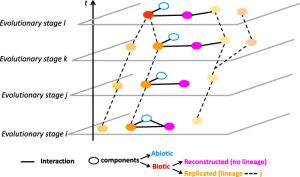
20. Bapteste E, Huneman P. Towards a Dynamic Interaction Network of Life to unify and expand the evolutionary theory. BMC Biol. 2018 May 29;16(1):56. doi: 10.1186/s12915-018-0531-6.
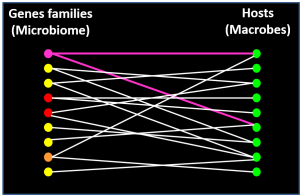
19.Vigliotti C, Bicep C, Bapteste E, Lopez P, Corel E. Tracking the Rules of Transmission and Introgression with Networks. Microbiol Spectr. 2018 Apr;6(2). doi: 10.1128/microbiolspec.MTBP-0008-2016.
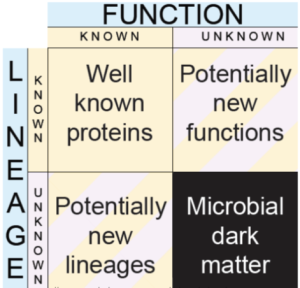
18. Bernard G., Pathmanathan J.S., Lannes R, Lopez L, Bapteste E. Microbial dark matter investigations: how microbial studies transform biological knowledge and empirically sketch a logic of scientific discovery, Genome Biology and Evolution. 2018. Feb 5.doi.org/10.1093/gbe/evy031
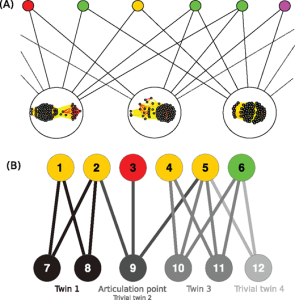
17. Corel E, Méheust R, Watson AK, McInerney JO, Lopez P, Bapteste E. Bipartite network analysis of gene sharings in the microbial world. Mol Biol Evol. 2018 Jan 15. doi: 10.1093/molbev/msy001.
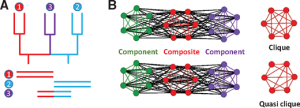
16. Pathmanathan JS, Lopez P, Lapointe FJ, Bapteste E. CompositeSearch: A Generalized Network Approach for Composite Gene Families Detection. Mol Biol Evol. 2018 Jan 1;35(1):252-255. doi: 10.1093/molbev/msx283.
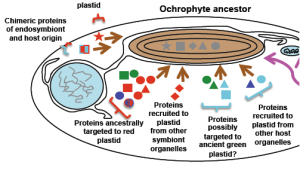
15. Richard G Dorrell, Gillian Gile, Giselle McCallum, Raphaël Méheust, Eric P Bapteste,Christen M Klinger, Loraine Brillet-Guéguen, Katalina D Freeman, Daniel J Richter, Chris Bowler (2017): Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife;10.7554/eLife.23717
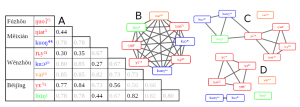
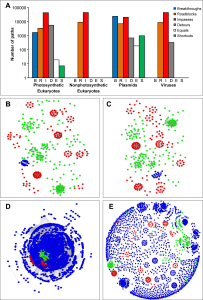
13. Lord E, Le Cam M, Bapteste É, Méheust R, Makarenkov V, Lapointe FJ. BRIDES: A New Fast Algorithm and Software for Characterizing Evolving Similarity Networks Using Breakthroughs, Roadblocks, Impasses, Detours, Equals and Shortcuts. PLoS One. 2016 Aug 31;11(8):e0161474.
12. List JM, Pathmanathan JS, Lopez P, Bapteste E. Unity and disunity in evolutionary sciences: process-based analogies open common research avenues for biology and linguistics. Biol Direct. 2016 Aug 20;11:39. doi: 10.1186/s13062-016-0145-2.
11. Jaffe AL, Corel E, Sylvestre Pathmanathan J, Lopez P, Bapteste E. Bipartite graph analyses reveal interdomain LGT involving ultrasmall prokaryotes and their divergent, membrane-related proteins. Environ Microbiol. 2016 Aug 3. doi: 10.1111/1462-2920.13477. [Epub ahead of print]
10. Méheust R, Zelzion E, Bhattacharya D, Lopez P, Bapteste E. Protein networks identify novel symbiogenetic genes resulting from plastid endosymbiosis. Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):3579-84. doi: 10.1073/pnas.1517551113. Epub 2016 Mar 14.
9. Corel E, Lopez P, Méheust R, Bapteste E. (2016) Network-Thinking: Graphs to Analyze Microbial Complexity and Evolution. Trends Microbiol. Jan 13. pii: S0966-842X(15)00279-6. doi: 10.1016/j.tim.2015.12.003. [Epub ahead of print]
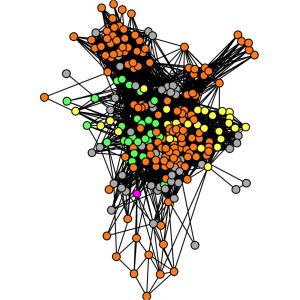
8. Lopez P, Halary S, Bapteste E. (2015) Highly divergent ancient gene families in metagenomic samples are compatible with additional divisions of life. Biology Direct. Oct 26; 10:64

7. Bapteste E. (2015) Conflits Intérieurs. Fable Scientifique, Collection « Essais », Les éditions Matériologiques (268 pages).
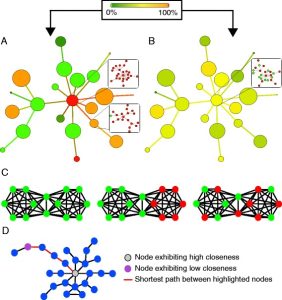
6. Forster D, Bittner L, Karkar S, Dunthorn M, Romac S, Audic S, Lopez P, Stoeck T, Bapteste E. (2015) Testing ecological theories with sequence similarity networks: marine ciliates exhibit similar geographic dispersal patterns as multicellular organisms. BMC Biol. Feb 24;13(1):16.
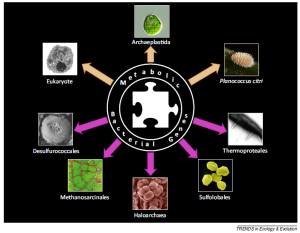
5. Méheust R, Lopez P and Bapteste E. (2015) Metabolic bacterial genes and the construction of high-level composite lineages of life. Trends Ecol Evol. Jan 16. pii: S0169-5347(15)00003-8. doi: 10.1016/j.tree.2015.01.001. [Epub ahead of print]
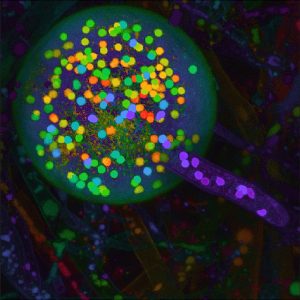
4. Boon E, Halary S, Bapteste E* and Hijri M*. (2015) Studying Genome Heterogeneity within the Arbuscular Mycorrhizal Fungal Cytoplasm. Genome Biol Evol. Jan 7. pii: evv002. [Epub ahead of print (* contributed equally to this work)

3. Bapteste E. (2014) Pluralisme et évolution réticulée en microbiologie évolutive in PRÉCIS DE PHILOSOPHIE DE LA BIOLOGIE, sous la direction de F. Merlin et T. Hoquet, Collection « Philosophie des sciences » dirigée par Thierry Martin, Vuibert (321 pages).
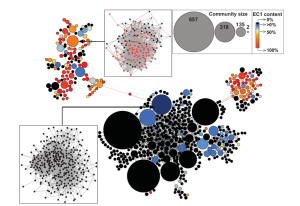
2. Cheng S., Karkar S., Bapteste E., Yee N., Falkowski P. and Bhattacharya D. Sequence similarity network reveals the imprints of major diversification events in the evolution of microbial life. Front. Ecol. Evol. 2014. doi: 10.3389/fevo.2014.00072
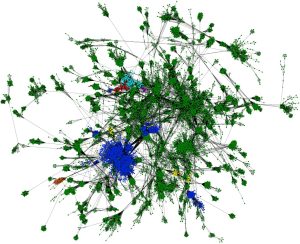
1. Jachiet P, Colson P, Lopez P, Bapteste E. Extensive gene remodeling in the viral world : new evidence for non-gradual evolution in the mobilome network. Genome Biol Evol. 2014;6(9):2195-205
*EVOLUNET SEMINARS*
15/04/2019: Workshop on networks and evolution
Dr. Eric Bapteste (ISYEB, CNRS, ERC PI, UPMC, 7 Quai saint Bernard, 75005 Paris, France) : “The networks of evolution”
Dr. Mary O’Connell (U. Nottingham, UK): ‘Gene remodeling and protein-coding innovation in the animal kingdom’
25/06/2018-30/06/2018: ‘Talks of the 2nd International Summer School on network and evolution’
Invited speakers :
Dr. Eric Bapteste (IBPS, CNRS, ERC PI, UPMC, 7 Quai saint Bernard, 75005 Paris, France) : “Introduction to sequence similarity networks”
Pr. James McInerney (University of Nottingham, UK): “Networks and public goods : reticulate evolution in prokaryotes”
Dr. Philippe Gambette (Université Paris-Est Marne-la-Vallée, LIGM, France): “The different types of networks used in evolutionary study : how they are made and what they are useful for”
Pr. Debashish Bhattacharya (Rutgers University, USA): “Reticulate evolution in eukaryotes”
Pr. Michel Habib (LIAFA, INRIA, France): “Graph centralities and graph comparisons”
Dr. Damien Eveillard (Université de Nantes, France): “Co-occurrence networks and the evolution of geochemical cycles in the environment”
Pr. Tal Dagan (Kiel University, Germany): “Directed bipartite graphs and LGT mediated by phages”
Pr. Mehdi Layeghifard (Toronto University, Canada): “Metagenomic networks”
Pr. Marc-André Sélosse (Institut de Systématique, Évolution, Biodiversité (ISYEB – UMR 7205 – CNRS, MNHN, UPMC, EPHE), Muséum national d’Histoire naturelle, Sorbonne Universités, 57 rue Cuvier, CP50, 75005, Paris, France): “The living world as a network”
22/02/2018: Dr. Michael Macey (The Open University, UK) gave a talk on: Analogues and Simulants – Investigations into the potential of life outside of Earth.
To know more about Dr. Macey’s work, please check: https://uk.linkedin.com/in/michael-macey-29881685

24/10/2017: ‘International Symposium on Network and Evolution’ (UPMC)
Invited speakers :
Dr. Christophe Dessimoz (U. Lausanne, Switzerland) : ‘Integrating genomes via fine-grained orthology’
Dr. Andrew Watson (UPMC, France): ‘Symbiogenetic genes in Haloarchaea’
Pr. Mark Ragan (U. Queensland, Brisbane, Australia): ‘Phylogenetic trees and networks without multiple sequence alignment’
Pr. Tal Dagan (U. Kiel, Germany) : ‘Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer via transduction’
Dr. Mattis List (U. Jena , Germany): ‘Using networks to infer sound-correspondence patterns across multiple languages’
09/12/2016: Pr. Eugene Koonin (Senior Investigator, NCBI, USA) gave a talk on « Evolution of the virus world and antivirus defense: a tangled web »
Find more about the work by Pr. Koonin here: https://www.ncbi.nlm.nih.gov/research/groups/koonin/
04/07/2016-08/07/2016: ‘Talks of the 1rst International Summer School on network and evolution’
Invited speakers :
Pr. Fernando Baquero, Val Fernández-Lanza (Group of Biology and Evolution of Microorganisms, IRYCIS, Department of Microbiology, Ramón y Cajal University Hospital, CIBERESP, Spain): “Reticulate Nested Evolution in the Clinical Bacterial World”
Pr. Robert G. Beiko (Faculty of Computer Science, Dalhousie University): “Lateral gene transfer: how many stories can a network tell?”
Pr. Marc-André Sélosse (Institut de Systématique, Évolution, Biodiversité (ISYEB – UMR 7205 – CNRS, MNHN, UPMC, EPHE), Muséum national d’Histoire naturelle, Sorbonne Universités, 57 rue Cuvier, CP50, 75005, Paris, France): “The evolution of interdependency by neutral evolution in holobionts”
Dr. E. Bapteste (CNRS, UPMC, France): “Network-Thinking: Graphs to Analyze Microbial Complexity and Evolution”
Dr. Marco Fondi (Dep. of Biology, University of Florence): “Microbial sequence similarity networks: deciphering the role of plasmids in microbial evolution and ecology.”
Pr. François-Joseph Lapointe (Département de sciences biologiques, Université de Montréal): “A statistical framework to assess and compare complex biological networks”
Dr. Mattis List (CRLAO and UPMC, France): “ Non-Tree-Like Processes in Language Evolution”
Dr. Philippe Huneman (IHPST, France): “Network analysis and the philosophy of scientific explanations.”
Pr. Michel Habib (LIAFA, INRIA, France): “Modeling with graphs, from success stories to quicksands”
29/04/2016: Pr. Martin Embley (Institute for Cell and Molecular Biosciences, Newcastle University, England) gave a talk on « The tangled origin of eukaryotes »
Find more about the work by Pr. Embley here: http://www.ncl.ac.uk/camb/staff/profile/martinembley.html#research
08/04/2015: Pr. Christopher Lane (U. Rhodes Island, USA) gave a talk on: « Falling in and out of parasitism: What can genomic data tell us about life history transitions? »
Find more about the work by Pr. Lane here: http://cels.uri.edu/bio/lanelab/
18/03/2015: Pr. James McInerney (U. Maynooth, Ireland) gave a talk on: « Eukaryotic origins »
Find more about the work by Pr. Mcinerney here: http://jamesmcinerney.ie/

24/11/2014: Pr. William Martin (U. Duesseldorf, Germany) gave a public talk on: »Major gene fluxes in early microbial evolution »
Find more about the work by Pr. Martin here: http://www.molevol.de/lab/martin.html
07/10/2014: Dr. Rafal Mostowy (Imperial College, UK) gave a public talk on: « The role of horizontal DNA exchange in the evolution of pneumococcal capsules ».
Find more about the work by Dr. Mostowy here: http://rmostowy.wordpress.com/

16/06/2014: Pr. Debashish Bhattacharya (Rutgers University, USA) gave a public talk on : « Exploring the evolution of microbial eukaryotes using single cell genomics ».
A challenge for network studies: exploiting data from single cell genomics to better understand protist ecology and evolution.
Find more exciting scientific contributions by Pr. Debashish Bhattacharya here: http://dblab.rutgers.edu/home/index.php
*SOFTWARES*
Composite-Search
MultiTwin
* DATA FROM THE EVOLUNET PROJECT*
Coming later 😉
*EUROPEAN CLASSES ON SEQUENCE SIMILARITY NETWORKS *
We plan to teach and share our methods on sequence similarity networks during two one-week European classes, each open to 10 motivated European master and PhD students in Station Biologique de Roscoff (France).
First class occurred in July 2016.
Second class occurred in June 2018.
Third class will occurr in June 2019.
If interested, apply to: eric.bapteste[at]snv.jussieu.fr
